Heddle Lab

DNA gyrase is a fascinating molecular nanomachine. It is an enzyme with the unique ability to negatively supercoil DNA. ~One of its roles is to remove torsion accumulated ahead of the replication fork during DNA replication. To achieve this, gyrase transiently introduces a double strand cleavage in its substrate DNA to facilitate the passage of one strand of DNA through another. This complex and highly coordinated mechanism of action has been studied for many years and recent advances in structural and biophysical techniques have allowed important details about the structure and function of gyrase to be unveiled. Not only does this bring us closer to understanding exactly how gyrase acts as a natural nanomachine, it may also provide us with key insights into how we might go about designing entirely artificial nanomachines for commercial or therapeutic applications.


Another feature of gyrase which is of particular therapeutic interest is that it is ubiquitous and essential in prokaryotes for maintenance of the bacterial chromosome but is not found in humans. This has been exploited by the widely used class of broad-spectrum antibacterial drug known as the fluoroquinolones which inhibit re-ligation of the cleaved DNA during the gyrase catalytic cycle, eventually leading to accumulation of double strand breaks which are lethal to the cell. However, the emergence of bacterial resistance against these drugs is of significant clinical concern. Understanding how this resistance arises is essential to either rescue the activity of drugs such as fluoroquinolones, or to aid the design of new antimicrobials.
Gyrase can also be found in some eukaryotes and is localised to subcellular organelles, as a result of endosymbiosis. One such example is the DNA gyrase found in the plant chloroplast required to maintain chloroplast DNA topology. Of important interest are the gyrases found in the apicoplast, a plastid within Apicomplexa. These gyrases have been identified as crucial for the survival of parasites such as those that cause malaria (Plasmodium falciparum) and toxoplasmosis (Toxoplasma gondii). However, so far it has not been possible to fully characterise these proteins and therefore, more research is required.

Gyrase publications:
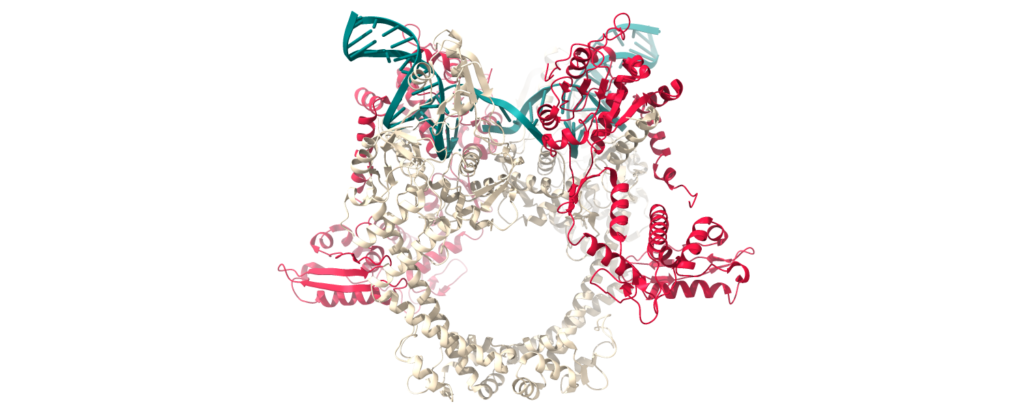
Michalczyk, E., Pakosz-Stepień, Z., Liston, J., Ghilarov, D., Gittins, O., Pabis, M., Heddle, J., Ghilarov, D. Structural basis of chiral wrap and T-segment capture by Escherichia coli DNA gyrase.
In: PNAS. 2024
PDF download
Saathoff M, Kosol S, Semmler T, Tedin K, Dimos N, Kupke J, Seidel M, Ghazisaeedi F, Jonske MC, Wolf SA, Kuropka B, Czyszczoń W, Ghilarov D, Grätz S, Heddle JG, Loll B, Süssmuth RD, Fulde M. Gene amplifications cause high-level resistance against albicidin in gram-negative bacteria.
In: PLoS Biol. 2023
PDF download
Michalczyk E, Hommernick K, Behroz I, Kulike M, Pakosz-Stępień Z, Mazurek L, Seidel M, Kunert M, Santos K, von Moeller H, Loll B, Weston JB, Mainz A, Heddle JG, Süssmuth RD, Ghilarov D. Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
In: Nat Catal. 2023
PDF download
Pakosz, Z., Lin, T. Y., Michalczyk, E., Nagano, S., Heddle, J. G.* (2021).Inhibitory compounds targeting Plasmodium falciparum gyrase B.,
In: Antimicrob. Agents Chemother.
PDF download
Mazurek, L., Ghilarov, D.*, Michalczyk, E., Pakosz, Z. Metelev, M., Czyszczon, W., Wawro, K., Behroz, I., Dubiley, S., Süssmuth, R., Heddle, J. G.* (2021) Pentapeptide repeat protein QnrB1 requires ATP hydrolysis to rejuvenate poisoned gyrase complexes.
In: Nucl. Acids Res.
PDF download
Nagano, S., Seki, E., Lin, T-Y., Shirouzu, M., Yokoyama, S., Heddle, J. G. (2015) Investigating the roles of the C-terminal domain of Plasmodium falciparum GyrA.
In: PLOS ONE
PDF download
Lin, T.-Y., Nagano, S. & Gardiner Heddle, J. (2015). Functional Analyses of the Toxoplasma gondii DNA Gyrase Holoenzyme: A Janus Topoisomerase with Supercoiling and Decatenation Abilities.
In: Sci. Rep.
PDF download



